

主管:中国科学院
主办:中国科学院古脊椎动物与古人类研究所
出版:科学出版社
主办:中国科学院古脊椎动物与古人类研究所
出版:科学出版社


人类学学报 ›› 2019, Vol. 38 ›› Issue (04): 491-498.doi: 10.16359/j.cnki.cn11-1963/q.2019.0068cstr: 32091.14.j.cnki.cn11-1963/q.2019.0068
收稿日期:2019-03-18
修回日期:2019-06-16
出版日期:2019-11-15
发布日期:2020-09-10
通讯作者:
黄石
作者简介:张野(1987-),男,博士,主要从事古DNA与人类进化研究。E-mail: 基金资助:Received:2019-03-18
Revised:2019-06-16
Online:2019-11-15
Published:2020-09-10
Contact:
HUANG Shi
摘要:
1983年,科学家们根据线粒体DNA(mtDNA)系统发育树构建了首个现代人起源的分子模型,认为现代人起源于亚洲,但1987年非洲起源说的提出取代了这一亚洲起源说。非洲起源说所依赖的无限多位点假说以及分子钟假说后来被普遍认为是错误的且不切实际的。我们近几年提出了一个新的分子进化模式,即遗传多样性上限理论,重新构建了一个新的人类起源模型。这一模型与多地区起源说基本吻合, 重新把现代人类起源地定位在了东亚。非洲说与东亚说在线粒体进化树上的主要区别是单倍型N和R的关系,非洲起源说认为N是R的祖先,东亚说则反之。本研究引用了已发表的古代人群mtDNA数据,重点分析了线粒体单倍群N和R的关系。结果显示,三个最古老的人类(一个距今45000年,其他两个约40000年)都属于单倍群R;在距今39500到30000年前的人类样本中,绝大部分属于单倍群R下游的亚单倍群U,只有两例为单倍群N(Oase1距今39500年,Salkhit距今34425年)。这两例所属单倍型位于单倍群N下游最基本的未分化亚型,不属于今天存在的任何N下游单倍型,所以可能靠近单倍群N的根部。这些古DNA数据揭示单倍群R比单倍群N古老大约5000年,进一步证实了亚洲起源说的正确性,非洲说的依据不足。
中图分类号:
张野, 黄石. 古DNA的新发现支持现代人东亚起源说[J]. 人类学学报, 2019, 38(04): 491-498.
ZHANG Ye, HUANG Shi. The Out of East Asia theory of modern human origins supported by recent ancient mtDNA findings[J]. Acta Anthropologica Sinica, 2019, 38(04): 491-498.
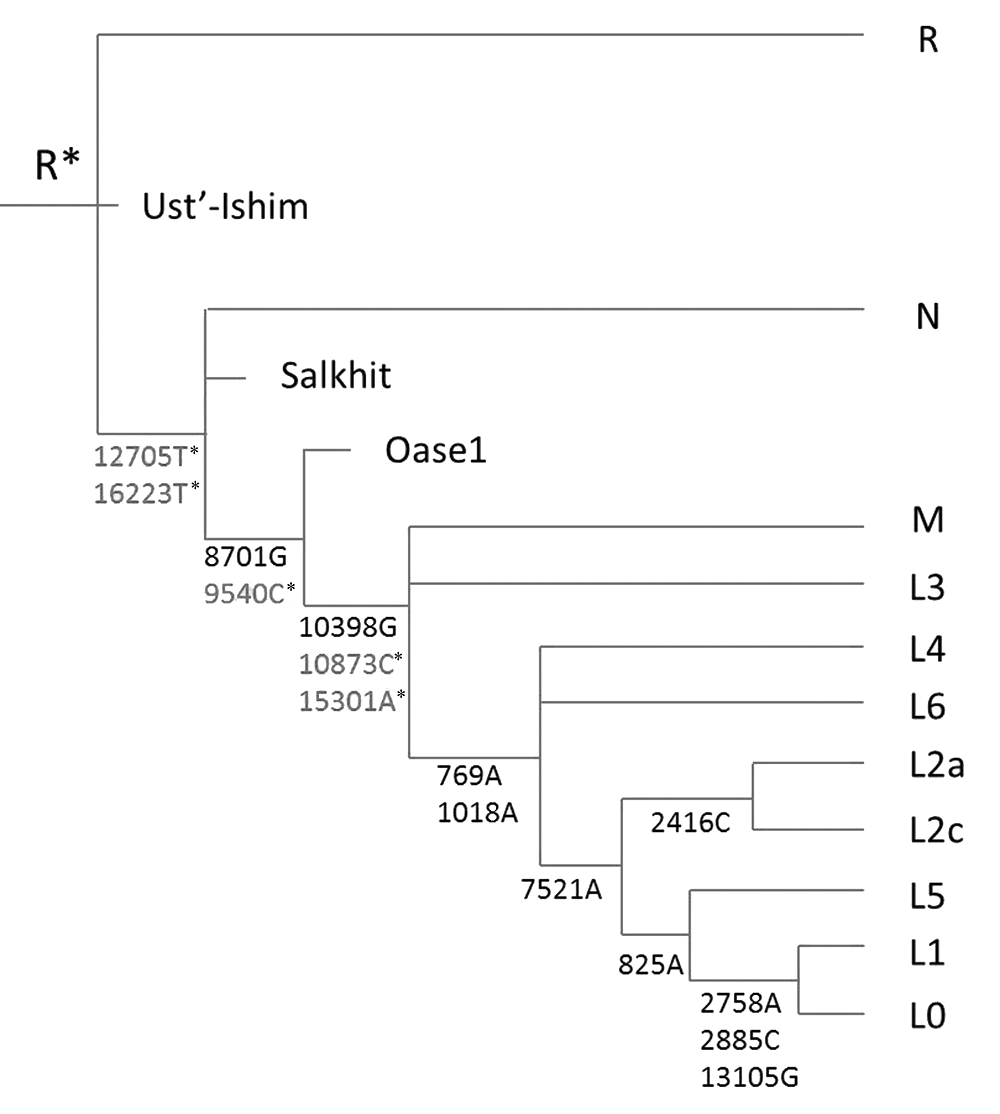
图1 出亚洲说线粒体系统发育树 只列出主要的单倍型分支以及代表性突变位点。 慢变位点(改变蛋白和RNA序列)标为正常黑体,快变位点黑体右上带有小星号“*”。同样列出了部分具有代表性的古人DNA单倍型。这些古DNA的位置是由他们的变异位点决定的。需要说明的是,本线粒体树的构建没有用到古代样本的信息, 完全是来自对今天单倍群的分析。
Fig.1 Phylogenetic tree of mtDNA according to the Out of East Asia model
| [1] |
Stringer CB, Andrews P. Genetic and fossil evidence for the origin of modern humans[J]. Science, 1988,239(4845):1263-1268
URL pmid: 3125610 |
| [2] |
Thorne AG, Wolpoff MH. Regional continuity in Australasian Pleistocene hominid evolution[J]. Am J Phys Anthropol, 1981,55:337-349
URL pmid: 6791505 |
| [3] | Wolpoff MH, Wu XZ, Thorne AG. Modern homo sapiens origins: a general theory of hominid evolution involving the fossil evidence from east Asia[M]. New York: Alan R. Liss, 1984 |
| [4] | Wu X. On the origin of modern humans in China[J]. Quaternary International, 2004,117:131-140 |
| [5] |
Yuan D, Lei X, Gui Y, et al. Modern human origins: multiregional evolution of autosomes and East Asia origin of Y and mtDNA[J]. bioRxiv, 2017: doi: https://doi.org/10.1101/106864
doi: 10.1101/2020.08.16.253013 URL pmid: 32839778 |
| [6] |
Cann RL, Stoneking AC, Wilson AC. Mitochondrial DNA and human evolution[J]. Nature, 1987,325:31-36
URL pmid: 3025745 |
| [7] |
Green RE, Krause J, Et., et al. A draft sequence of the Neandertal Genome[J]. Science, 2010,328:710-722
URL pmid: 20448178 |
| [8] |
Meyer M, Kircher M, Gansauge MT, et al. A High-Coverage Genome Sequence from an Archaic Denisovan Individual[J]. Science, 2012
URL pmid: 32883851 |
| [9] |
Fu Q, Hajdinjak M, Moldovan OT, et al. An early modern human from Romania with a recent Neanderthal ancestor[J]. Nature, 2015,524(7564):216-219
URL pmid: 26098372 |
| [10] |
Fu Q, Li H, Moorjani P, et al. Genome sequence of a 45,000-year-old modern human from western Siberia[J]. Nature, 2014,514(7523):445-449
doi: 10.1038/nature13810 URL pmid: 25341783 |
| [11] |
Vernot B, Akey JM. Resurrecting surviving Neandertal lineages from modern human genomes[J]. Science, 2014,343(6174):1017-21
doi: 10.1126/science.1245938 URL pmid: 24476670 |
| [12] |
Hublin JJ, Ben-Ncer A, Bailey SE, et al. New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens[J]. Nature, 2017,546(7657):289-292
URL pmid: 28593953 |
| [13] |
White TD, Asfaw B, Degusta D, et al. Pleistocene Homo sapiens from Middle Awash, Ethiopia[J]. Nature, 2003,423:742-747
doi: 10.1038/nature01669 URL pmid: 12802332 |
| [14] |
Scerri EML, Thomas MG, Manica A, et al. Did Our Species Evolve in Subdivided Populations across Africa, and Why Does It Matter?[J]. Trends Ecol Evol, 2018,33(8):582-594
doi: 10.1016/j.tree.2018.05.005 URL pmid: 30007846 |
| [15] |
Liu W, Martinon-Torres M, Cai YJ, et al. The earliest unequivocally modern humans in southern China[J]. Nature, 2015,526(7575):696-699
doi: 10.1038/nature15696 URL pmid: 26466566 |
| [16] |
Johnson MJ, Wallace DC, Ferris SD, et al. Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns[J]. J Mol Evol, 1983,19(3-4):255-271
URL pmid: 6310133 |
| [17] |
Ayala FJ. Molecular clock mirages[J]. BioEssays, 1999,21(1):71-75
doi: 10.1002/(SICI)1521-1878(199901)21:1<71::AID-BIES9>3.0.CO;2-B URL pmid: 10070256 |
| [18] |
Ayala FJ. On the virtues and pitfalls of the molecular evolutionary clock[J]. J Hered, 1986,77(4):226-235
URL pmid: 3020121 |
| [19] |
Hu T, Long M, Yuan D, et al. The genetic equidistance result, misreading by the molecular clock and neutral theory and reinterpretation nearly half of a century later[J]. Sci China Life Sci, 2013,56:254-261
URL pmid: 23526392 |
| [20] | Huang S. The genetic equidistance result of molecular evolution is independent of mutation rates[J]. J. Comp. Sci. Syst. Biol. , 2008,1:92-102 |
| [21] | Huang S. The overlap feature of the genetic equidistance result, a fundamental biological phenomenon overlooked for nearly half of a century[J]. Biological Theory, 2010,5:40-52 |
| [22] |
Kumar S. Molecular clocks: four decades of evolution[J]. Nat Rev Genet, 2005,6(8):654-662
URL pmid: 16136655 |
| [23] |
Hahn MW. Toward a selection theory of molecular evolution[J]. Evolution, 2008,62(2):255-265
URL pmid: 18302709 |
| [24] |
Kreitman MW. The neutral theory is dead. Long live the neutral theory[J]. Bioessays, 1996, 18(8): 678-83; discussion 683
URL pmid: 8760341 |
| [25] |
Ohta T, Gillespie JH. Development of Neutral and Nearly Neutral Theories[J]. Theor Popul Biol, 1996,49(2):128-142
URL pmid: 8813019 |
| [26] |
Ballard JW, Kreitman M. Is mitochondrial DNA a strictly neutral marker?[J]. Trends Ecol Evol, 1995,10(12):485-488
doi: 10.1016/s0169-5347(00)89195-8 URL pmid: 21237113 |
| [27] | Leffler EM, Bullaughey K, Matute DR, et al. Revisiting an old riddle: what determines genetic diversity levels within species?[J]. PLoS Biol, 2012,10(9):e1001388 |
| [28] |
Huang S. New thoughts on an old riddle: What determines genetic diversity within and between species?[J]. Genomics, 2016,108(1):3-10
URL pmid: 26835965 |
| [29] |
Zhu Z, Lu Q, Wang J, et al. Collective effects of common SNPs in foraging decisions in Caenorhabditis elegans and an integrative method of identification of candidate genes[J]. Sci. Rep., 2015: doi: 10.1038/srep16904
URL pmid: 32895444 |
| [30] |
Zhu Z, Yuan D, Luo D, et al. Enrichment of Minor Alleles of Common SNPs and Improved Risk Prediction for Parkinson’s Disease[J]. PLoS ONE, 2015,10(7):e0133421
URL pmid: 26207627 |
| [31] |
Zhu Z, Man X, Xia M, et al. Collective effects of SNPs on transgenerational inheritance in Caenorhabditis elegans and budding yeast[J]. Genomics, 2015,106(1):23-9
doi: 10.1016/j.ygeno.2015.04.002 URL pmid: 25882787 |
| [32] |
Biswas K, Chakraborty S, Podder S, et al. Insights into the dN/dS ratio heterogeneity between brain specific genes and widely expressed genes in species of different complexity[J]. Genomics, 2016,108(1):11-7
URL pmid: 27126306 |
| [33] |
Zhu Z, Lu Q, Zeng F, et al. Compatibility between mitochondrial and nuclear genomes correlates wtih quantitative trait of lifespan in Caenorhabditis elegans[J]. Sci. Rep., 2015: doi: 10.1038/srep17303
URL pmid: 32895444 |
| [34] |
Huang S. Primate phylogeny: molecular evidence for a pongid clade excluding humans and a prosimian clade containing tarsiers[J]. Sci China Life Sci, 2012,55:709-725
doi: 10.1007/s11427-012-4350-7 URL pmid: 22932887 |
| [35] | Huang S. Inverse relationship between genetic diversity and epigenetic complexity[J]. Preprint available at Nature Precedings 2009: doi.org/10.1038/npre.2009.1751.2 |
| [36] | Huang S. Histone methylation and the initiation of cancer, Cancer Epigenetics[M]. New York: CRC Press, 2008 |
| [37] |
Margoliash E. Primary structure and evolution of cytochrome c[J]. Proc. Natl. Acad. Sci. , 1963,50:672-679
doi: 10.1073/pnas.50.4.672 URL pmid: 14077496 |
| [38] |
Luo D, Huang S. The genetic equidistance phenomenon at the proteomic level[J]. Genomics, 2016,108(1):25-30
URL pmid: 26973320 |
| [39] |
Yuan D, Huang S. On the peopling of the Americas: molecular evidence for the Paleoamerican and the Solutrean models[J]. bioRxiv, 2017, bioRxiv 130989; doi: https://doi.org/10.1101/130989
doi: 10.1101/2020.08.16.253013 URL pmid: 32839778 |
| [40] |
Zhang Y, Lei X, Chen H, et al. Ancient DNAs and the Neolithic Chinese super-grandfather Y haplotypes[J]. bioRxiv, 2018, doi: https://doi.org/10.1101/487918
URL pmid: 32839778 |
| [41] |
Meyer M, Fu Q, Aximu-Petri A, et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos[J]. Nature, 2014,505(7483):403-406
doi: 10.1038/nature12788 URL pmid: 24305051 |
| [42] |
Teske P R, Golla T R, Sandoval-Castillo J, et al. Mitochondrial DNA is unsuitable to test for isolation by distance[J]. Sci Rep, 2018,8(1):8448
doi: 10.1038/s41598-018-25138-9 URL pmid: 29855482 |
| [43] |
Towarnicki SG, Ballard JWO. Mitotype Interacts With Diet to Influence Longevity, Fitness, and Mitochondrial Functions in Adult Female Drosophila[J]. Front Genet, 2018,9:593
doi: 10.3389/fgene.2018.00593 URL pmid: 30555517 |
| [44] |
Fu Q, Meyer M, Gao X, et al. DNA analysis of an early modern human from Tianyuan Cave, China[J]. Proc Natl Acad Sci U S A, 2013,110(6):2223-2227
doi: 10.1073/pnas.1221359110 URL pmid: 23341637 |
| [45] |
Benazzi S, Slon V, Talamo S, et al. Archaeology. The makers of the Protoaurignacian and implications for Neandertal extinction[J]. Science, 2015,348(6236):793-796
URL pmid: 25908660 |
| [46] |
Deviese T, Massilani D, Yi S, et al. Compound-specific radiocarbon dating and mitochondrial DNA analysis of the Pleistocene hominin from Salkhit Mongolia[J]. Nat Commun, 2019,10(1):274
URL pmid: 30700710 |
| [47] |
Hershkovitz I, Weber GW, Quam R, et al. The earliest modern humans outside Africa[J]. Science, 2018,359(6374):456-459
URL pmid: 29371468 |
| [48] |
Groucutt H S, Grun R, Zalmout I a S, et al. Homo sapiens in Arabia by 85,000 years ago[J]. Nat Ecol Evol, 2018,2(5):800-809
doi: 10.1038/s41559-018-0518-2 URL pmid: 29632352 |
| [49] |
Athreya S, Wu X. A multivariate assessment of the Dali hominin cranium from China: Morphological affinities and implications for Pleistocene evolution in East Asia[J]. Am J Phys Anthropol, 2017,164(4):679-701
URL pmid: 29068047 |
| [50] | Zhao L, Zhang L, Du B, et al. New discovery of human fossils and associated mammal faunas in Bijie Guizhou[J]. Acta Anthropologica Sinica, 2016,35:24-35 |
| [51] | Curnoe D, Ji X, Shajin H, et al. Dental remains from Longtanshan cave 1 (Yunnan, China), and the initial presence of anatomically modern humans in East Asia[J]. Quaternary International, 2016,400:180-186 |
| [52] | Bae CJ, Wang W, Zhao J, et al. Modern human teeth from Late Pleistocene Luna Cave(Guangxi, China)[J]. Quaternary International, 2014,354:169-183 |
| [53] |
Li Z Y, Wu XJ, Zhou LP, et al. Late Pleistocene archaic human crania from Xuchang, China[J]. Science, 2017,355(6328):969-972
doi: 10.1126/science.aal2482 URL pmid: 28254945 |
| [54] |
Fu Q, Posth C, Hajdinjak M, et al. The genetic history of Ice Age Europe[J]. Nature, 2016,534(7606):200-205
URL pmid: 27135931 |
| [55] |
Posth C, Renaud G, Mittnik A, et al. Pleistocene Mitochondrial Genomes Suggest a Single Major Dispersal of Non-Africans and a Late Glacial Population Turnover in Europe[J]. Curr Biol, 2016,26(6):827-833
URL pmid: 26853362 |
| [1] | 张明, 平婉菁, YANG Melinda Anna, 付巧妹. 古基因组揭示史前欧亚大陆现代人复杂遗传历史[J]. 人类学学报, 2023, 42(03): 412-421. |
| [2] | 丁曼雨, 何伟, 王恬怡, 夏格旺堆, 张明, 曹鹏, 刘峰, 戴情燕, 付巧妹. 中国西藏拉托唐古墓地古代居民线粒体全基因组研究[J]. 人类学学报, 2021, 40(01): 1-11. |
| [3] | 王恬怡, 赵东月, 张明, 乔诗雨, 杨帆, 万杨, 杨若薇, 曹鹏, 刘峰, 付巧妹. 古DNA捕获新技术与中国南方早期人群遗传研究新格局[J]. 人类学学报, 2020, 39(04): 680-694. |
| [4] | 赵静, 王传超. 古DNA提取技术对比及概述[J]. 人类学学报, 2020, 39(04): 706-716. |
| [5] | 李春香, 张帆, 马鹏程, 王立新, 崔银秋. 线粒体全基因组揭示嫩江流域史前人群遗传结构的动态变化[J]. 人类学学报, 2020, 39(04): 695-705. |
| [6] | 张雅军, 张旭, 赵欣, 仝涛, 李林辉. 从头骨形态学和古DNA探究公元3~4世纪西藏阿里地区人群的来源[J]. 人类学学报, 2020, 39(03): 435-449. |
| [7] | 张明;付巧妹. 史前古人类之间的基因交流及对当今现代人的影响[J]. 人类学学报, 2018, 37(02): 206-218. |
| [8] | 雷晓云;袁德健;张野;黄石. 基于DNA分子的现代人起源研究35年回顾与展望[J]. 人类学学报, 2018, 37(02): 270-283. |
| [9] | 文少卿;王传超;敖雪;韦兰海;佟欣竹;王凌翔;王占峰;韩昇;李辉. 古DNA证据支持曹操的父系遗传类型属于单倍群O2[J]. 人类学学报, 2016, 35(04): 617-625. |
| [10] | 张芃胤; 徐智; 许渤松; 韩康信; 周慧; 金力; 谭婧泽. 青海大通上孙家寨古代居民mtDNA遗传分析[J]. 人类学学报, 2013, 32(02): 204-218. |
| [11] | 冉鹏;吴谨;张蓓蕾;单华鑫;李英碧. 人类线粒体DNA异质性研究进展及其法医学应用[J]. 人类学学报, 2007, 26(04): 372-378. |
| [12] | 李彬彬;钟复光;易红生;王先然;李良芳;王丽兰;齐晓岚;吴立甫. 贵州四个民族人群线粒体DNA RegionⅤ的遗传多态性[J]. 人类学学报, 2006, 25(03): 258-262. |
| [13] | 赵凌霞;王翠斌. 安达曼岛民的起源问题[J]. 人类学学报, 2006, 25(02): 105-105. |
| [14] | 何惠琴,金建中,许淳,姜言睿,朱琪泉,谭婧泽,黄微,徐永庆,金力,任大明. 3200年前中国新疆哈密古人骨的mtDNA多态性研究[J]. 人类学学报, 2003, 22(04): 329-337. |
| [15] | 杨东亚. 古代DNA研究中污染的控制和识别[J]. 人类学学报, 2003, 22(02): 163-173. |
| 阅读次数 | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
全文 1155
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
摘要 3130
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
 京ICP证05002819号-3
京ICP证05002819号-3