

主管:中国科学院
主办:中国科学院古脊椎动物与古人类研究所
出版:科学出版社
主办:中国科学院古脊椎动物与古人类研究所
出版:科学出版社


人类学学报 ›› 2020, Vol. 39 ›› Issue (04): 680-694.doi: 10.16359/j.cnki.cn11-1963/q.2020.0059cstr: 32091.14.j.cnki.cn11-1963/q.2020.0059
王恬怡1,2,3( ), 赵东月1,*(
), 赵东月1,*( ), 张明2,3,4, 乔诗雨2,3,4, 杨帆5, 万杨5, 杨若薇2,3, 曹鹏2,3, 刘峰2,3, 付巧妹2,3,4,*(
), 张明2,3,4, 乔诗雨2,3,4, 杨帆5, 万杨5, 杨若薇2,3, 曹鹏2,3, 刘峰2,3, 付巧妹2,3,4,*( )
)
收稿日期:2020-07-27
修回日期:2020-10-10
出版日期:2020-11-15
发布日期:2020-11-06
通讯作者:
赵东月,付巧妹
作者简介:王恬怡(1996-),女,云南个旧人,西北大学文化遗产学院硕士研究生,主要从事古DNA研究。Email:基金资助:
WANG Tianyi1,2,3( ), ZHAO Dongyue1,*(
), ZHAO Dongyue1,*( ), ZHANG Ming2,3,4, QIAO Shiyu2,3,4, YANG Fan5, WAN Yang5, YANG Ruowei2,3, CAO Peng2,3, LIU Feng2,3, FU Qiaomei2,3,4,*(
), ZHANG Ming2,3,4, QIAO Shiyu2,3,4, YANG Fan5, WAN Yang5, YANG Ruowei2,3, CAO Peng2,3, LIU Feng2,3, FU Qiaomei2,3,4,*( )
)
Received:2020-07-27
Revised:2020-10-10
Online:2020-11-15
Published:2020-11-06
Contact:
ZHAO Dongyue,FU Qiaomei
摘要:
古DNA捕获技术目前已获得极大的发展,能够从骨骼和环境沉积物等多种材料中获取到目的DNA片段,而且对于保存环境较差的低纬度地区,同样能够获取有效的内源DNA片段,极大地丰富了古DNA研究的材料来源。本文围绕这一新技术开展总结和讨论,主要分为两个方面:1)总结并介绍该技术应用前景;2)应用这一技术打开了中国南方早期人群研究的新局面,梳理该新技术的应用对史前中国南方人群古基因组研究所获得的新认识,并对中国南方早期人群古基因组深入分析;另外,利用古DNA捕获技术成功获取云南3446~3180 BP的大阴洞遗址4例高质量线粒体基因组信息,并开展了人群遗传历史的研究。
中图分类号:
王恬怡, 赵东月, 张明, 乔诗雨, 杨帆, 万杨, 杨若薇, 曹鹏, 刘峰, 付巧妹. 古DNA捕获新技术与中国南方早期人群遗传研究新格局[J]. 人类学学报, 2020, 39(04): 680-694.
WANG Tianyi, ZHAO Dongyue, ZHANG Ming, QIAO Shiyu, YANG Fan, WAN Yang, YANG Ruowei, CAO Peng, LIU Feng, FU Qiaomei. Ancient DNA capture techniques and genetic study progress of early southern China populations[J]. Acta Anthropologica Sinica, 2020, 39(04): 680-694.
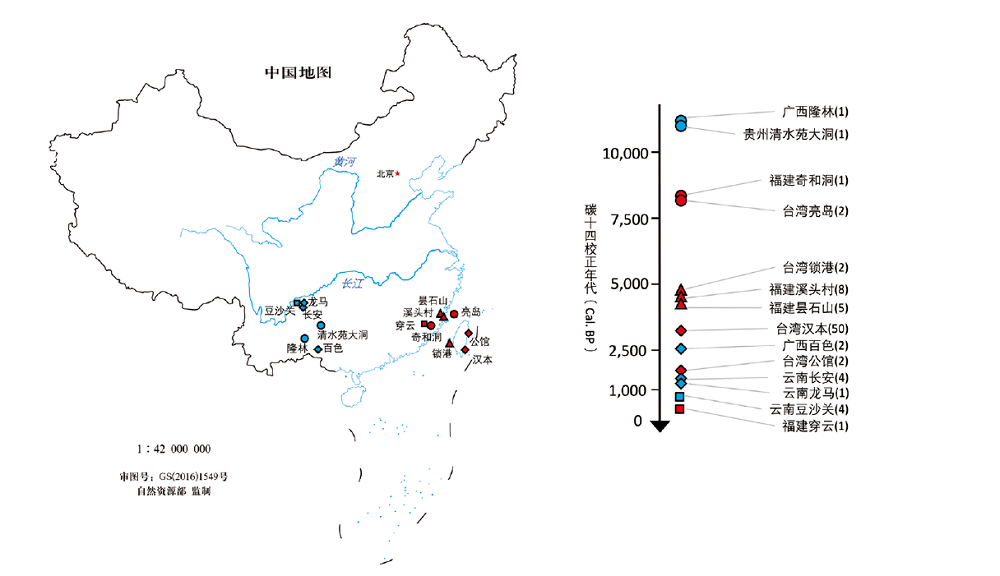
图2 中国南方已发表的古DNA样本信息 不同颜色表示所发表古DNA数据的类型:红色表示核基因组数据,蓝色表示线粒体基因组数据。不同形状代表了不同的年代:圆形表示个体碳测年在距今7500年以前,三角形表示距今7500到4000年之间的个体,菱形表示距今4000年到距今1000年的个体,正方形表示距今1000年以内的个体。括号中的数字代表样本数量。
Fig.2 Information on published ancient DNA samples in southern China Red indicates the data is the nuclear genome, blue indicates it’s the mitochondrial genome. The different symbol means different time periods. The sample sizes of sequenced individuals are put in the brackets.
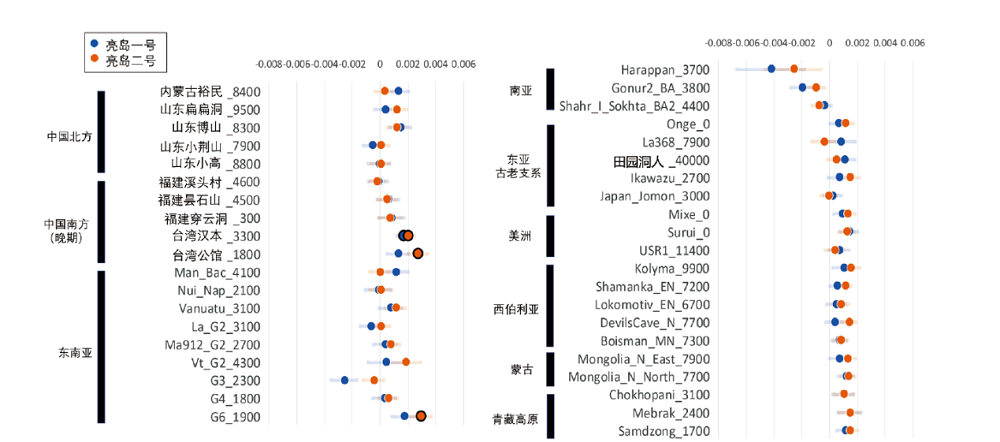
图3 f4-statistics分析图。 蓝色表示f4(Mbuti,X;Qihe,Liangdao1),橙色表示f4(Mbuti,X;Qihe,Liangdao2),每个点的数值对应人群计算所得的f4值,浅色线段表示误差线即f4±std.err。f4-statistics具有显著性的结果(|Z|>3)圆点加了黑色边缘。若f4值小于零,即f4(Mbuti,X;Qihe,Liangdao1/Liangdao2)<0,表示该人群相较于两个亮岛人与奇和洞人的遗传关系更近,大于零则反之。样本名称后为样本距今年代
Fig.3 The result of f4(Mbuti, X; Qihe, Liangdao1/Liangdao2), where X is various ancient populations from East Eurasia. The blue indicates the results of f4(Mbuti, X; Qihe, Liangdao1). The orange indicates the results of f4(Mbuti, X; Qihe, Liangdao2). Circle with black edge are significant results with |Z|>3. If the f4-value lesser than 0, suggesting the “X” population is closer to Qihe than Liangdao. The number after the sample name is the date before present.
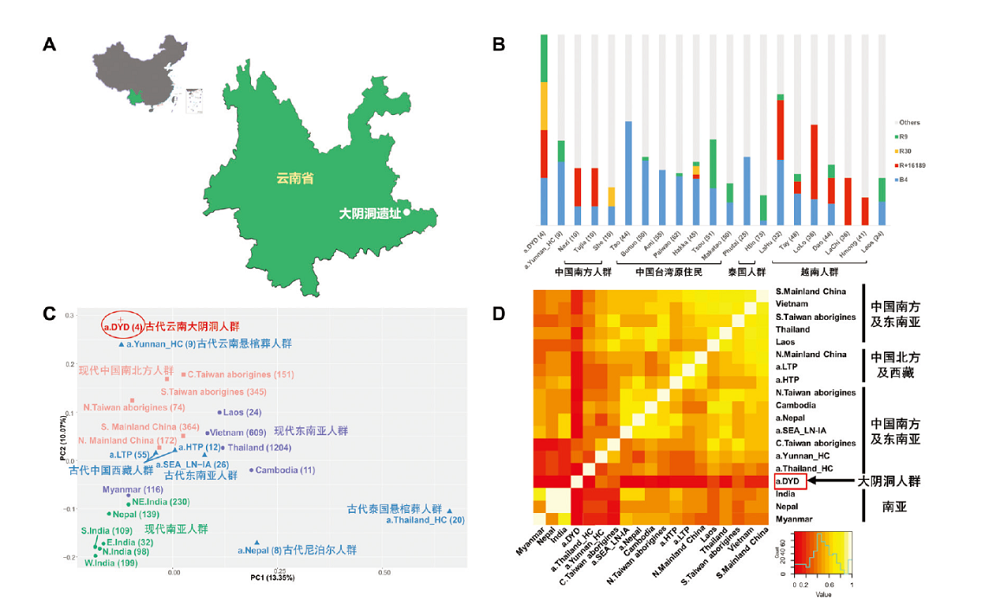
图4 大阴洞地理信息以及线粒体基因组分析结果 A) 大阴洞遗址地理位置location of Dayin Cave site;B) 大阴洞个体相关单倍群分布直方图histogram of haplogroup distributions;C) 基于单倍群频率的主成分分析图PCA analysis based on the haplogroup frequency;D) 共享单倍群值热图heatmap of haplogroup sharing calculation。图中大阴洞古代个体以a.DYD表示,括号中的数字表示本研究中该人群样本的数量。
Fig.4 Geographic information and mtDNA genome analyses of Dayin Cave The ancient individuals of Dayin Cave are represented by “a.DYD”, and each number in the bracket shows the sample size from the population in this study.
| 样本Sample ID | 取样部位 Skeletal Element | 线粒体DNA 平均覆盖度 Mean Coverage of mtDNA | 平均污染度% Contamination of mtDNA% | 污染度% (95%置信区间) Contamination% (95%CI) | 单倍型 Haplotype | 14C校正年代 Calibrated years (cal BP) |
|---|---|---|---|---|---|---|
| 2017GDM10 | 指骨 | 108.23 | 2.0 | 0.9-4.1 | R+16189 | 3446-3326 |
| 2017GDM6:R1 | 距骨 | 226.34 | 1.6 | 0.8-2.4 | R9c1b1 | 3360-3180 |
| 2017GDM1 | 距骨 | 217.98 | 1.1 | 0.5-2.1 | R30 | 3367-3213 |
| 2017GDM7:R1 | 指骨 | 308.30 | 1.9 | 1.2-2.7 | B4c1b2a | 3444-3249 |
| 2017GDM12* | 尺骨 | 10.13 | 10.7 | 6.6-16.7 | H+195 | 3386-3237 |
表1 大阴洞样本信息
Tab.1 Information of the Dayin Cave samples
| 样本Sample ID | 取样部位 Skeletal Element | 线粒体DNA 平均覆盖度 Mean Coverage of mtDNA | 平均污染度% Contamination of mtDNA% | 污染度% (95%置信区间) Contamination% (95%CI) | 单倍型 Haplotype | 14C校正年代 Calibrated years (cal BP) |
|---|---|---|---|---|---|---|
| 2017GDM10 | 指骨 | 108.23 | 2.0 | 0.9-4.1 | R+16189 | 3446-3326 |
| 2017GDM6:R1 | 距骨 | 226.34 | 1.6 | 0.8-2.4 | R9c1b1 | 3360-3180 |
| 2017GDM1 | 距骨 | 217.98 | 1.1 | 0.5-2.1 | R30 | 3367-3213 |
| 2017GDM7:R1 | 指骨 | 308.30 | 1.9 | 1.2-2.7 | B4c1b2a | 3444-3249 |
| 2017GDM12* | 尺骨 | 10.13 | 10.7 | 6.6-16.7 | H+195 | 3386-3237 |
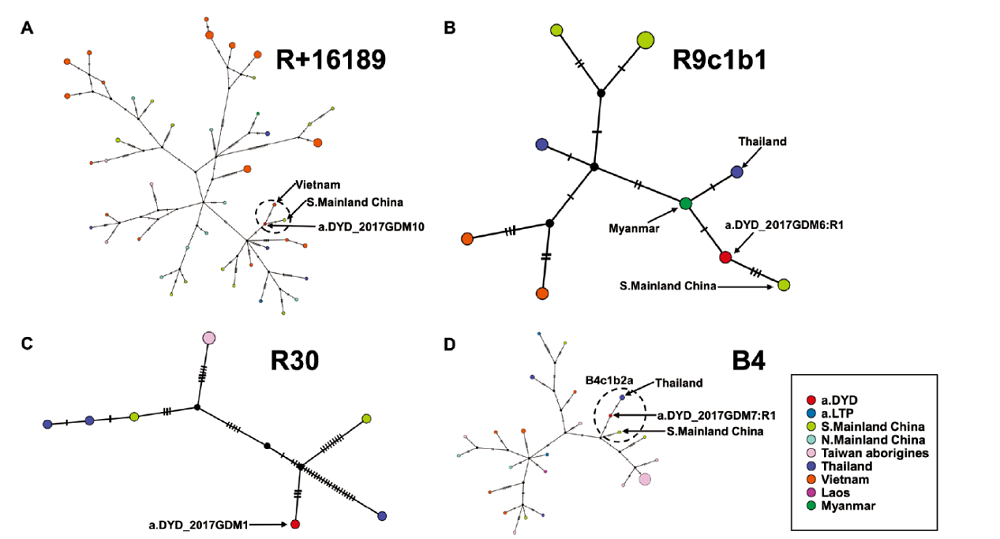
图5 大阴洞个体所属单倍型网络结构图 A) R+16189单倍型网络结构图network of R+16189;B) R9c1b1单倍型网络结构图network of R9c1b1;C) R30单倍型网络结构图network of R30;D) B4单倍型网络结构图network of B4。箭头指出大阴洞个体以及与大阴洞个体所属单倍型相关的人群。
Fig.5 Haplotype networks of individuals from Dayin Cave Black arrows indicate the Dayin Cave individuals and the populations associated with haplotypes which Dayin Cave individuals belong to.
| [1] |
Yang MA, Fan X, Sun B, et al. Ancient DNA indicates human population shifts and admixture in northern and southern China[J]. Science, 2020,369(6501):282-288
URL pmid: 32409524 |
| [2] |
Bai F, Zhang X, Ji X, et al. Paleolithic genetic link between Southern China and Mainland Southeast Asia revealed by ancient mitochondrial genomes[J]. Journal of Human Genetics, 2020: 1-4
doi: 10.1007/s100380050001 URL pmid: 10697955 |
| [3] | Ko AMS, Chen CY, Fu QM, et al. Early Austronesians: into and out of Taiwan[J]. American Journal of Human Genetics, 2014,94(3):426-436 |
| [4] | Zhang X, Li C, Zhou Y, et al. A Matrilineal Genetic Perspective of Hanging Coffin Custom in Southern China and Northern Thailand[J]. iScience, 2020,23(4):101032 |
| [5] | Wang CC, Yeh HY, Popov AN, et al. The Genomic Formation of Human Populations in East Asia[J]. bioRxiv, 2020 |
| [6] | Yang MA, Gao X, Theunert C, et al. 40,000-year-old individual from Asia provides insight into early population structure in Eurasia[J]. Current Biology, 2017, 27(20): 3202-3208.e9 |
| [7] | Ko AMS, Zhang Y, Yang MA, et al. Mitochondrial genome of a 22,000-year-old giant panda from southern China reveals a new panda lineage[J]. Current Biology, 2018,28(12):R693-R694 |
| [8] | Slon V, Hopfe C, Weiss CL, et al. Neandertal and Denisovan DNA from Pleistocene sediments[J]. Science, 2017,356(6338):605-608 |
| [9] |
Burbano HA, Green RE, Maricic T, et al. Analysis of human accelerated DNA regions using archaic hominin genomes[J]. PloS one, 2012,7(3):e32877
doi: 10.1371/journal.pone.0032877 URL pmid: 22412940 |
| [10] | Burbano HA, Hodges E, Green RE, et al. Targeted investigation of the Neandertal genome by array-based sequence capture[J]. Science, 2010,328(5979):723-725 |
| [11] | Ávila-Arcos MC, Cappellini E, Romero-Navarro JA, et al. Application and comparison of large-scale solution-based DNA capture-enrichment methods on ancient DNA[J]. Scientific reports, 2011,1(1):1-5 |
| [12] | Maricic T, Whitten M, Pääbo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products[J]. PloS one, 2010,5(11):e14004 |
| [13] |
Sheng GL, Basler N, Ji XP, et al. Paleogenome reveals genetic contribution of extinct giant panda to extant populations[J]. Current Biology, 2019, 29(10): 1695-1700. e6
URL pmid: 31080081 |
| [14] | Fu QM, Meyer M, Gao X, et al. DNA analysis of an early modern human from Tianyuan Cave, China[J]. Proceedings of the National Academy of Sciences, 2013,110(6):2223-2227 |
| [15] |
Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing[J]. Nature biotechnology, 2009,27(2):182-189
doi: 10.1038/nbt.1523 URL pmid: 19182786 |
| [16] |
Fu QM, Hajdinjak M, Moldovan O T, et al. An early modern human from Romania with a recent Neanderthal ancestor[J]. Nature, 2015,524(7564):216-219
URL pmid: 26098372 |
| [17] |
Fu QM, Li H, Moorjani P, et al. Genome sequence of a 45,000-year-old modern human from western Siberia[J]. Nature, 2014,514(7523):445-449
URL pmid: 25341783 |
| [18] | Fu QM, Posth C, Hajdinjak M, et al. The genetic history of ice age Europe[J]. Nature, 2016,534(7606):200-205 |
| [19] | Kistler L, Ware R, Smith O, et al. A new model for ancient DNA decay based on paleogenomic meta-analysis[J]. Nucleic Acids Research, 2017,45(11):6310-6320 |
| [20] |
Meyer M, Arsuaga JL, de Filippo C, et al. Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins[J]. Nature, 2016,531(7595):504-507
doi: 10.1038/nature17405 URL pmid: 26976447 |
| [21] | Gansauge MT, Meyer M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA[J]. Nature protocols, 2013,8(4):737-748 |
| [22] | Glocke I, Meyer M. Extending the spectrum of DNA sequences retrieved from ancient bones and teeth[J]. Genome research, 2017,27(7):1230-1237 |
| [23] |
Meyer M, Kircher M, Gansauge MT, et al. A high-coverage genome sequence from an archaic Denisovan individual[J]. Science, 2012,338(6104):222-226
doi: 10.1126/science.1224344 URL pmid: 22936568 |
| [24] |
Lipson M, Skoglund P, Spriggs M, et al. Population turnover in Remote Oceania shortly after initial settlement[J]. Current Biology, 2018, 28(7): 1157-1165.e7
doi: 10.1016/j.cub.2018.02.051 URL pmid: 29501328 |
| [25] | McColl H, Racimo F, Vinner L, et al. The prehistoric peopling of Southeast Asia[J]. Science, 2018,361(6397):88-92 |
| [26] | Zhang M, Sun G, Ren L, et al. Ancient DNA evidence from China reveals the expansion of Pacific dogs[J]. Molecular Biology and Evolution, 2020,37(5):1462-1469 |
| [27] |
de Barros Damgaard P, Marchi N, Rasmussen S, et al. 137 ancient human genomes from across the Eurasian steppes[J]. Nature, 2018,557(7705):369-374
URL pmid: 29743675 |
| [28] |
de Barros Damgaard P, Martiniano R, Kamm J, et al. The first horse herders and the impact of early Bronze Age steppe expansions into Asia[J]. Science, 2018,360(6396)
doi: 10.1126/science.360.6396.1391 URL pmid: 29954963 |
| [29] |
Ning C, Wang CC, Gao S, et al. Ancient genomes reveal Yamnaya-related ancestry and a potential source of indo-European speakers in iron age Tianshan[J]. Current Biology, 2019, 29(15): 2526-2532.e4
URL pmid: 31353181 |
| [30] |
Sikora M, Pitulko VV, Sousa VC, et al. The population history of northeastern Siberia since the Pleistocene[J]. Nature, 2019,570(7760):182-188
URL pmid: 31168093 |
| [31] |
Siska V, Jones ER, Jeon S, et al. Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago[J]. Science Advances, 2017,3(2):e1601877
URL pmid: 28164156 |
| [32] | Wong EHM, Khrunin A, Nichols L, et al. Reconstructing genetic history of Siberian and Northeastern European populations[J]. Genome research, 2017,27(1):1-14 |
| [33] | Pei S, Gao X, Wang H, et al. The Shuidonggou site complex: new excavations and implications for the earliest Late Paleolithic in North China[J]. Journal of Archaeological Science, 2012,39(12):3610-3626 |
| [34] | Wu X, Liu W, Wang Z. A human parietal fossil found at the Shuidonggou site, Ningxia, China[J]. Anthropological Science, 2004,112(1):83-89 |
| [35] | Yang S X, Deng C L, Zhu R X, et al. The Paleolithic in the Nihewan Basin, China: Evolutionary history of an Early to Late Pleistocene record in Eastern Asia[J]. Evolutionary Anthropology: Issues, News, and Reviews, 2020,29(3):125-142 |
| [36] | 高星. 更新世东亚人群连续演化的考古证据及相关问题论述[J]. 人类学学报, 2014,33(3):237-253 |
| [37] | Cai P, Huang Q, Zhang X, et al. Adsorption of DNA on clay minerals and various colloidal particles from an Alfisol[J]. Soil Biology and Biochemistry, 2006,38(3):471-476 |
| [38] | Ogram A, Sayler GS, Gustin D, et al. DNA adsorption to soils and sediments[J]. Environmental science & technology, 1988,22(8):982-984 |
| [39] |
Willerslev E, Hansen A J, Binladen J, et al. Diverse plant and animal genetic records from Holocene and Pleistocene sediments[J]. Science, 2003,300(5620):791-795
URL pmid: 12702808 |
| [40] | Nichols RV, Curd E, Heintzman PD, et al. Targeted Amplification and Sequencing of Ancient Environmental and Sedimentary DNA[J]. Methods in molecular biology (Clifton, NJ), 2019,1963:149-161 |
| [41] |
Pedersen MW, Ruter A, Schweger C, et al. Postglacial viability and colonization in North America’s ice-free corridor[J]. Nature, 2016,537(7618):45-49
URL pmid: 27509852 |
| [42] |
Matsumura H, Hung H, Higham C, et al. Craniometrics reveal “two layers” of prehistoric human dispersal in eastern Eurasia[J]. Scientific reports, 2019,9(1):1-12
doi: 10.1038/s41598-018-37186-2 URL pmid: 30626917 |
| [43] |
Westaway KE, Louys J, Awe RD, et al. An early modern human presence in Sumatra 73,000-63,000 years ago[J]. Nature, 2017,548(7667):322-325
URL pmid: 28792933 |
| [44] | Demeter F, Shackelford L, Westaway K, et al. Early modern humans from Tam Pà Ling, Laos: Fossil review and perspectives[J]. Current Anthropology, 2017,58(S17):S527-S538 |
| [45] | Matsumura H, Oxenham M. Population dispersal from East Asia into Southeast Asia: Evidence from cranial and dental Morphology[A]. In: Bioarchaeology of East Asia: Movement, Contact, Health[M]. University Press of Florida, 2013: 179-209 |
| [46] |
Lipson M, Cheronet O, Mallick S, et al. Ancient genomes document multiple waves of migration in Southeast Asian prehistory[J]. Science, 2018,361(6397):92-95
doi: 10.1126/science.aat3188 URL pmid: 29773666 |
| [47] | 焦天龙. 东南沿海的史前文化与南岛语族的扩散[J]. 中原文物, 2002 (2):13-16 |
| [48] | Blust RA. The proto-Austronesian pronouns and Austronesian subgrouping[J]. Working Papers in Linguistics, University of Hawaii, Honolulu, 1977,9:1-15 |
| [49] | Melton T, Peterson R, Redd AJ, et al. Polynesian genetic affinities with Southeast Asian populations as identified by mtDNA analysis[J]. American journal of human genetics, 1995,57(2):403-414 |
| [50] |
Raghavan M, Skoglund P, Graf KE, et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans[J]. Nature, 2014,505(7481):87-91
doi: 10.1038/nature12736 URL pmid: 24256729 |
| [51] | Diamond JM. Express train to Polynesia[J]. Nature, 1988,336(6197):307-308 |
| [52] | Jiao T. The Neolithic Archaeology of Southeast China[J]. A Companion to Chinese Archaeology, 2013: 597-611 |
| [53] | 张光直. 中国东南海岸考古与南岛语族起源问题[J]. 南方民族考古, 1987,1 |
| [54] | Chang K. Prehistoric and early historic culture horizons and traditions in South China[J]. Current Anthropology, 1964,5(5):359-375 |
| [55] | Chang K, Goodenough W H. Archaeology of southeastern coastal China and its bearing on the Austronesian homeland[J]. Transactions of the American philosophical society, 1996,86(5):36-56 |
| [56] | Bellwood P. Austronesian prehistory in Southeast Asia: homeland, expansion and transformation[M]. Canberra: ANU E Press, 1995 |
| [57] | 刘益昌. 台湾史前史专论 [M]. 台北: 联经出版事业股份有限公司, 2016 |
| [58] | 赵志军. 中国农业起源概述[J]. 遗产与保护研究, 2019 (1):1 |
| [59] | Crawford GW, Chen X, Luan F, et al. People and plant interaction at the Houli Culture Yuezhuang site in Shandong Province, China[J]. The Holocene, 2016,26(10):1594-1604 |
| [60] | Deng Z, Hung H, Fan X, et al. The ancient dispersal of millets in southern China: New archaeological evidence[J]. The Holocene, 2018,28(1):34-43 |
| [61] |
Jeong C, Ozga AT, Witonsky DB, et al. Long-term genetic stability and a high-altitude East Asian origin for the peoples of the high valleys of the Himalayan arc[J]. Proceedings of the National Academy of Sciences, 2016,113(27):7485-7490
doi: 10.1073/pnas.1520844113 URL |
| [62] |
Moreno-Mayar JV, Potter BA, Vinner L, et al. Terminal Pleistocene Alaskan genome reveals first founding population of Native Americans[J]. Nature, 2018,553(7687):203-207
doi: 10.1038/nature25173 URL pmid: 29323294 |
| [63] |
Shinde V, Narasimhan VM, Rohland N, et al. An ancient Harappan genome lacks ancestry from Steppe pastoralists or Iranian farmers[J]. Cell, 2019, 179(3): 729-735.e10
doi: 10.1016/j.cell.2019.08.048 URL pmid: 31495572 |
| [64] | Ding M, Wang T, Ko AMS, et al. Ancient mitogenomes show plateau populations from last 5200 years partially contributed to present-day Tibetans[J]. Proceedings of the Royal Society B, 2020,287(1923):20192968 |
| [65] |
Bhandari S, Zhang X, Cui C, et al. Genetic evidence of a recent Tibetan ancestry to Sherpas in the Himalayan region[J]. Scientific reports, 2015,5:16249
doi: 10.1038/srep16249 URL pmid: 26538459 |
| [66] |
Chandrasekar A, Kumar S, Sreenath J, et al. Updating phylogeny of mitochondrial DNA macrohaplogroup M in India: dispersal of modern human in South Asian corridor[J]. PloS one, 2009,4(10):e7447
doi: 10.1371/journal.pone.0007447 URL pmid: 19823670 |
| [67] |
Duong NT, Macholdt E, Ton ND, et al. Complete human mtDNA genome sequences from Vietnam and the phylogeography of Mainland Southeast Asia[J]. Scientific reports, 2018,8(1):1-13
doi: 10.1038/s41598-017-17765-5 URL pmid: 29311619 |
| [68] | Fornarino S, Pala M, Battaglia V, et al. Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation[J]. BMC Evolutionary Biology, 2009,9(1):154 |
| [69] |
Kang L, Zheng H X, Zhang M, et al. MtDNA analysis reveals enriched pathogenic mutations in Tibetan highlanders[J]. Scientific reports, 2016,6(1):1-11
doi: 10.1038/s41598-016-0001-8 URL pmid: 28442746 |
| [70] |
Kutanan W, Kampuansai J, Srikummool M, et al. Complete mitochondrial genomes of Thai and Lao populations indicate an ancient origin of Austroasiatic groups and demic diffusion in the spread of Tai-Kadai languages[J]. Human genetics, 2017,136(1):85-98
doi: 10.1007/s00439-016-1742-y URL pmid: 27837350 |
| [71] |
Li YC, Wang HW, Tian JY, et al. Ancient inland human dispersals from Myanmar into interior East Asia since the Late Pleistocene[J]. Scientific reports, 2015,5:9473
doi: 10.1038/srep09473 URL pmid: 25826227 |
| [72] | Lippold S, Xu H, Ko A, et al. Human paternal and maternal demographic histories: insights from high-resolution Y chromosome and mtDNA sequences[J]. Investigative genetics, 2014,5(1):13 |
| [73] | Peng MS, Palanichamy MG, Yao YG, et al. Inland post-glacial dispersal in East Asia revealed by mitochondrial haplogroup M9a'b[J]. BMC biology, 2011,9(1):2 |
| [74] |
Peng MS, Xu W, Song JJ, et al. Mitochondrial genomes uncover the maternal history of the Pamir populations[J]. European Journal of Human Genetics, 2018,26(1):124-136
doi: 10.1038/s41431-017-0028-8 URL pmid: 29187735 |
| [75] |
Qin Z, Yang Y, Kang L, et al. A mitochondrial revelation of early human migrations to the Tibetan Plateau before and after the last glacial maximum[J]. American Journal of Physical Anthropology, 2010,143(4):555-569
doi: 10.1002/ajpa.21350 URL pmid: 20623602 |
| [76] | Summerer M, Horst J, Erhart G, et al. Large-scale mitochondrial DNA analysis in Southeast Asia reveals evolutionary effects of cultural isolation in the multi-ethnic population of Myanmar[J]. BMC evolutionary biology, 2014,14(1):1-12 |
| [77] |
Wang HW, Li YC, Sun F, et al. Revisiting the role of the Himalayas in peopling Nepal: insights from mitochondrial genomes[J]. Journal of human genetics, 2012,57(4):228-234
doi: 10.1038/jhg.2012.8 URL pmid: 22437208 |
| [78] |
Renaud G, Stenzel U, Kelso J. leeHom: adaptor trimming and merging for Illumina sequencing reads[J]. Nucleic acids research, 2014,42(18):e141-e141
doi: 10.1093/nar/gku699 URL pmid: 25100869 |
| [79] |
Renaud G, Stenzel U, Maricic T, et al. deML: robust demultiplexing of Illumina sequences using a likelihood-based approach[J]. Bioinformatics, 2015,31(5):770-772
doi: 10.1093/bioinformatics/btu719 URL pmid: 25359895 |
| [80] |
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform[J]. bioinformatics, 2009,25(14):1754-1760
doi: 10.1093/bioinformatics/btp324 URL pmid: 19451168 |
| [81] |
Andrews RM, Kubacka I, Chinnery PF, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA[J]. Nature genetics, 1999,23(2):147
doi: 10.1038/13779 URL pmid: 10508508 |
| [82] |
Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools[J]. Bioinformatics, 2009,25(16):2078-2079
doi: 10.1093/bioinformatics/btp352 URL pmid: 19505943 |
| [83] |
Patterson N, Moorjani P, Luo Y, et al. Ancient admixture in human history[J]. Genetics, 2012,192(3):1065-1093
doi: 10.1534/genetics.112.145037 URL pmid: 22960212 |
| [84] |
Fu QM, Mittnik A, Johnson PLF, et al. A revised timescale for human evolution based on ancient mitochondrial genomes[J]. Current biology, 2013,23(7):553-559
doi: 10.1016/j.cub.2013.02.044 URL pmid: 23523248 |
| [85] | Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity[J]. BMC bioinformatics, 2004,5(1):113 |
| [86] |
Kloss-Brandstätter A, Pacher D, Schönherr S, et al. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups[J]. Human mutation, 2011,32(1):25-32
doi: 10.1002/humu.21382 URL pmid: 20960467 |
| [87] | Van Oven M. PhyloTree Build 17: Growing the human mitochondrial DNA tree[J]. Forensic Science International: Genetics Supplement Series, 2015,5:e392-e394 |
| [88] |
Van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation[J]. Human mutation, 2009,30(2):E386-E394
doi: 10.1002/humu.20921 URL pmid: 18853457 |
| [89] |
Bandelt H J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies[J]. Molecular biology and evolution, 1999,16(1):37-48
URL pmid: 10331250 |
| [90] | Leigh JW, Bryant D, Nakagawa S. PopART: Full-feature software for haplotype network construction. Methods in Ecology and Evolution, 2015,6(9):1110-1116 |
| [1] | 廖卫. 中国南方猩猩化石的研究进展[J]. 人类学学报, 2024, 43(02): 199-213. |
| [2] | 张明, 平婉菁, YANG Melinda Anna, 付巧妹. 古基因组揭示史前欧亚大陆现代人复杂遗传历史[J]. 人类学学报, 2023, 42(03): 412-421. |
| [3] | 浣发祥, 杨石霞, 陈苇, 陈伟驹, 朱利东, 张玉修. 四川汉源富林遗址石制品再研究[J]. 人类学学报, 2023, 42(02): 177-190. |
| [4] | 赵东月, 吕正, 张泽涛, 刘波, 凌雪, 万杨, 杨帆. 通过稳定同位素分析云南大阴洞遗址先民的生业经济方式[J]. 人类学学报, 2022, 41(02): 295-307. |
| [5] | 丁曼雨, 何伟, 王恬怡, 夏格旺堆, 张明, 曹鹏, 刘峰, 戴情燕, 付巧妹. 中国西藏拉托唐古墓地古代居民线粒体全基因组研究[J]. 人类学学报, 2021, 40(01): 1-11. |
| [6] | 李春香, 张帆, 马鹏程, 王立新, 崔银秋. 线粒体全基因组揭示嫩江流域史前人群遗传结构的动态变化[J]. 人类学学报, 2020, 39(04): 695-705. |
| [7] | 赵静, 王传超. 古DNA提取技术对比及概述[J]. 人类学学报, 2020, 39(04): 706-716. |
| [8] | 张雅军, 张旭, 赵欣, 仝涛, 李林辉. 从头骨形态学和古DNA探究公元3~4世纪西藏阿里地区人群的来源[J]. 人类学学报, 2020, 39(03): 435-449. |
| [9] | 张明;付巧妹. 史前古人类之间的基因交流及对当今现代人的影响[J]. 人类学学报, 2018, 37(02): 206-218. |
| [10] | Anne DAMBRICOURT MALASSé;ZHANG Pu;Patricia WILS. A new molar from the Middle Pleistocene hominid assemblage of Yanhuidong, Tongzi, South China[J]. 人类学学报, 2018, 37(01): 1-17. |
| [11] | 陈君;王頠;李大伟;廖卫. 广西田东中山遗址洞外岩厦出土动物骨骼的初步研究[J]. 人类学学报, 2017, 36(04): 527-536. |
| [12] | 文少卿;王传超;敖雪;韦兰海;佟欣竹;王凌翔;王占峰;韩昇;李辉. 古DNA证据支持曹操的父系遗传类型属于单倍群O2[J]. 人类学学报, 2016, 35(04): 617-625. |
| [13] | 张芃胤; 徐智; 许渤松; 韩康信; 周慧; 金力; 谭婧泽. 青海大通上孙家寨古代居民mtDNA遗传分析[J]. 人类学学报, 2013, 32(02): 204-218. |
| [14] | 陈少坤; 黄万波; 裴健; 贺存定; 秦利; 魏光飚; 冷静. 三峡地区最晚更新世的梅氏犀兼述中国南方更新世的犀牛化石[J]. 人类学学报, 2012, 31(04): 381-394. |
| [15] | 金昌柱;郑家坚;王元;徐钦琦. 中国南方早更新世主要哺乳动物群层序对比和动物地理[J]. 人类学学报, 2008, 27(04): 304-317. |
| 阅读次数 | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
全文 2237
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
摘要 1634
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
 京ICP证05002819号-3
京ICP证05002819号-3